Viewing Jupyter notebooks at the command line
The Jupyter notebook is a literate programming environment that has become ubiquitous in machine learning. While the standard tools for interacting with notebooks are web applications, it’s often useful to be able to view notebooks at the command line. This is convenient when logged into a training workstation via SSH, and the process of configuring SSH to forward a port, starting a Jupyter server, and navigating to it in a web browser is a chore to view a notebook for a few seconds.
Viewing other literate programming formats such as R Markdown and Pweave at the command line is trivial: these use lightweight markup languages that don’t need special rendering to be legible. As such it’s enough to print the raw file contents to the terminal, for example with cat.
This isn’t the case for Jupyter notebooks. A notebook is a structured JSON document conforming to a schema. Notebooks can contain images and other binary data, which if viewed with a pager will print unintelligible Base64 encoded data to your terminal. Characters such as " are backslash-escaped, making copying code from the output difficult.
Instead of printing the notebook file directly to the terminal, we’ll convert it to another format first. nbconvert is a program to convert notebooks to rich formats including HTML and PDF, as well as plain text such as Markdown. By design, Markdown syntax is unobtrusive and readable in plain text without rendering, so it’s a good choice for our output format.
As an example, we’ll use a notebook that contains both prose and code cells taken from the JAX documentation.
We use jupyter nbconvert to convert the notebook to Markdown, printing the output to standard output.
$ jupyter nbconvert --stdout --to markdown quickstart.ipynb
[NbConvertApp] Converting notebook quickstart.ipynb to markdown
# JAX Quickstart
**JAX is NumPy on the CPU, GPU, and TPU, with great automatic
differentiation for high-performance machine learning research.**
```python
import jax.numpy as jnp
from jax import grad, jit, vmap
from jax import random
```
## Multiplying Matrices
We'll be generating random data in the following examples. One big
difference between NumPy and JAX is how you generate random numbers. For
more details, see [Common Gotchas in JAX].
[Common Gotchas in JAX]: https://jax.readthedocs.io/en/latest/notebooks/Common_Gotchas_in_JAX.html#%F0%9F%94%AA-Random-Numbers
```python
key = random.PRNGKey(0)
x = random.normal(key, (10,))
print(x)
```
nbconvert writes the header [NbConvertApp] Converting notebook quickstart.ipynb to markdown to standard error and the notebook body to standard output, so we can filter out the header by redirecting standard error.
This redirection syntax is for Bourne-like shells such as Bash. If you’re using an alternative such as fish, change as appropriate.
This is already more readable than the raw JSON document, but we can improve the readability of the code cells by applying syntax highlighting. We’ll pipe the output of nbconvert to pygmentize, a command line interface to the Pygments syntax highlighting library. Pygments is a dependency of nbconvert, so if you’ve followed to here you’ll already have it installed.
As we’re piping text to pygmentize on standard input, there’s no filename from which to determine the language of the input so we specify it using the -l flag.
|
We pipe the output of pygmentize to a pager like less to scroll through and search within the notebook.
|
|
The -R flag instructs less to output ANSI escape sequences in raw form to colour the output. If we run this command, we’ll get a legible rendering of the notebook with syntax highlighting. Thanks to less, we can scroll up and down through the notebook, and search within it by pressing / and typing a query.
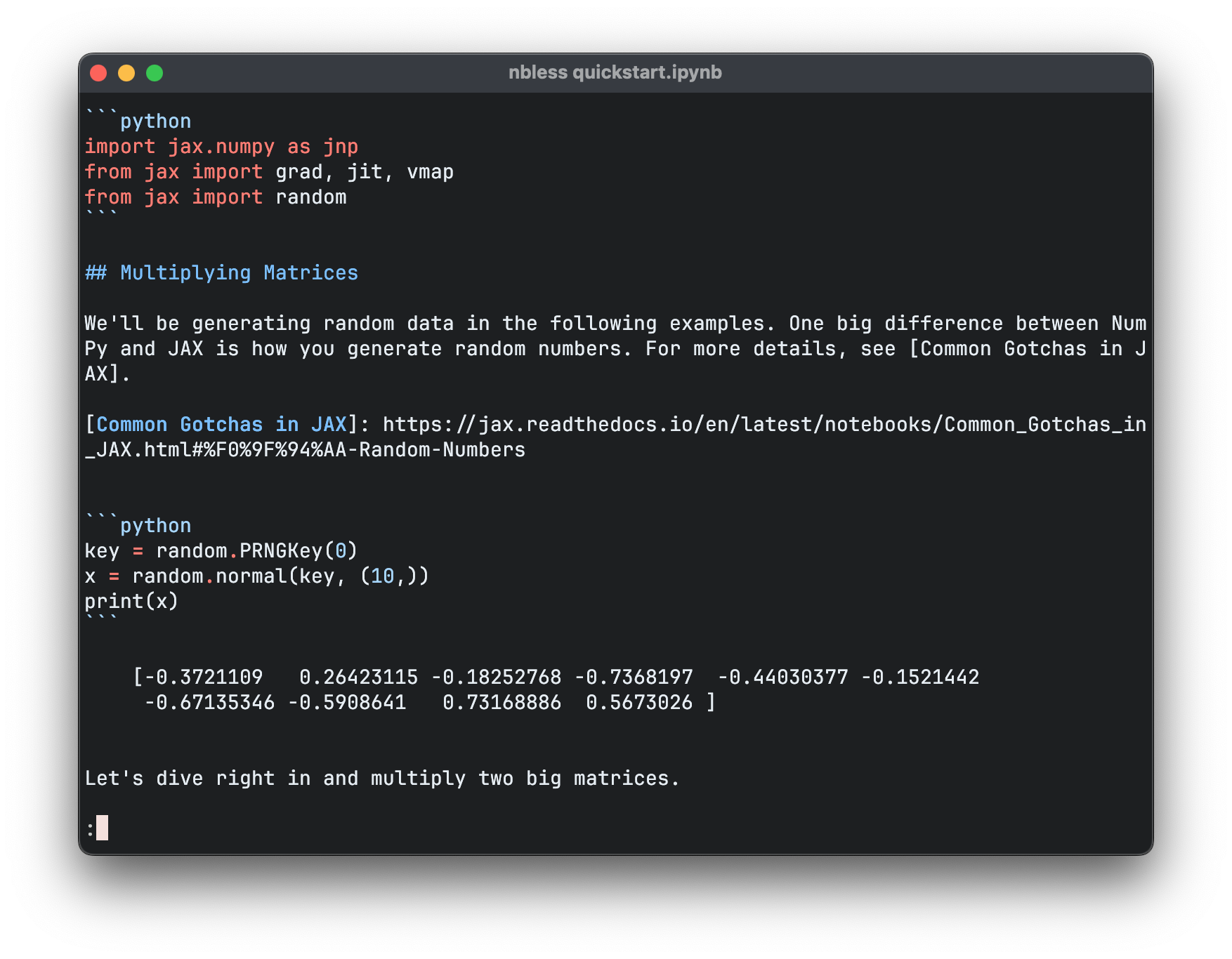
This is a longer command than is convenient to type for each use, so we create a shell script to encapsulate it, and also take the opportunity to add input validation.
#!/usr/bin/env bash
if ; then
fi
if ; then
fi
if ; then
fi
if ! ; then
fi
|
|
If we save this as an executable script on our shell’s command search path, for example as nbless, next time we’re at the command line on a remote machine and want to view a notebook we can run nbless notebook.ipynb–no port forwarding, Jupyter server, or web browser required.